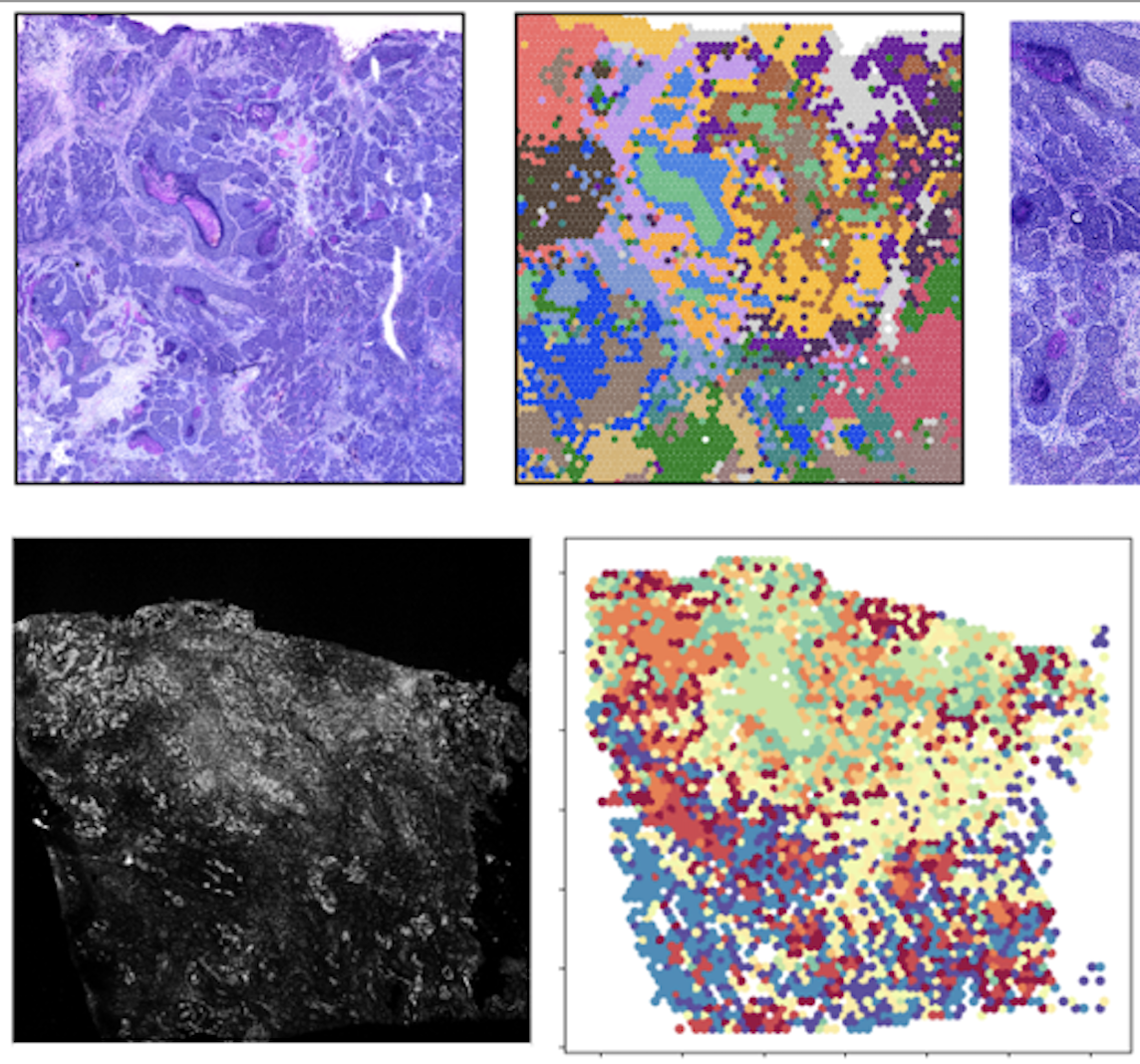
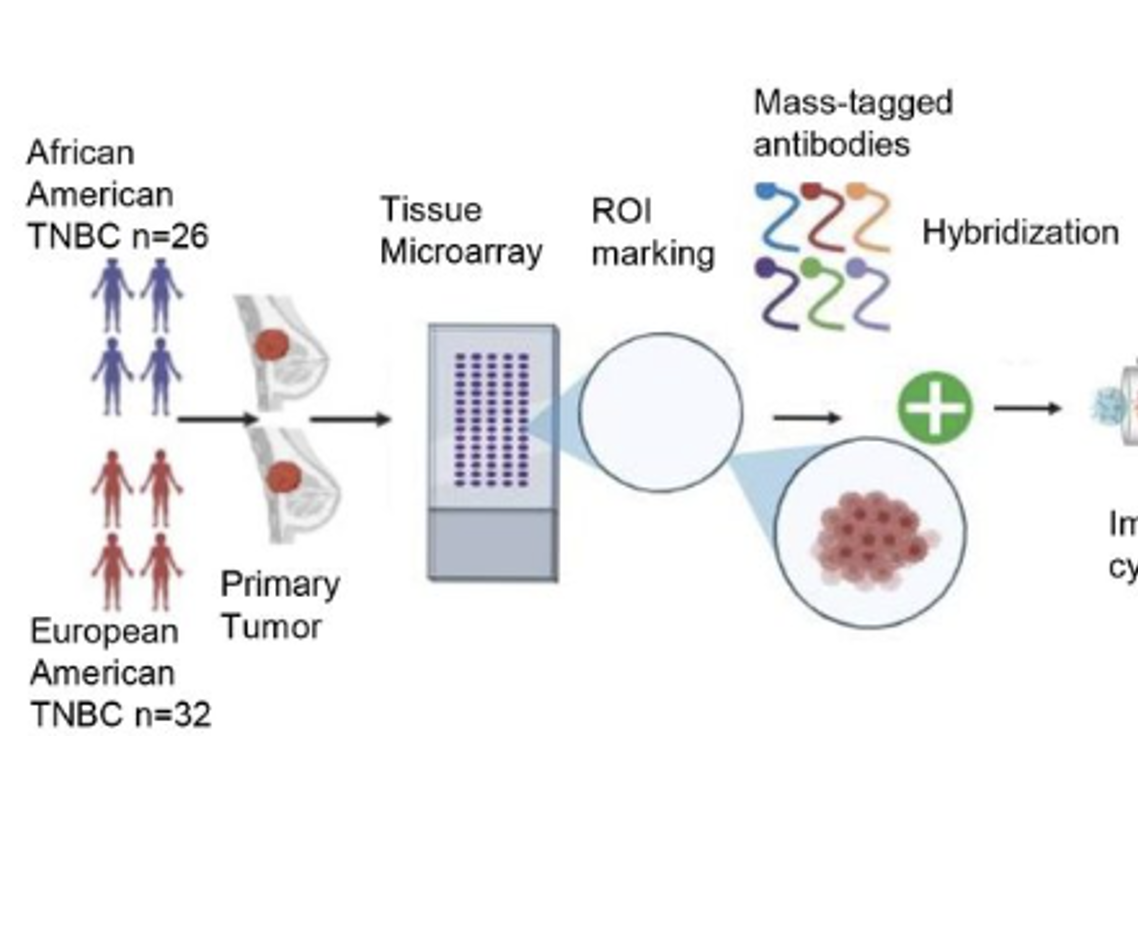
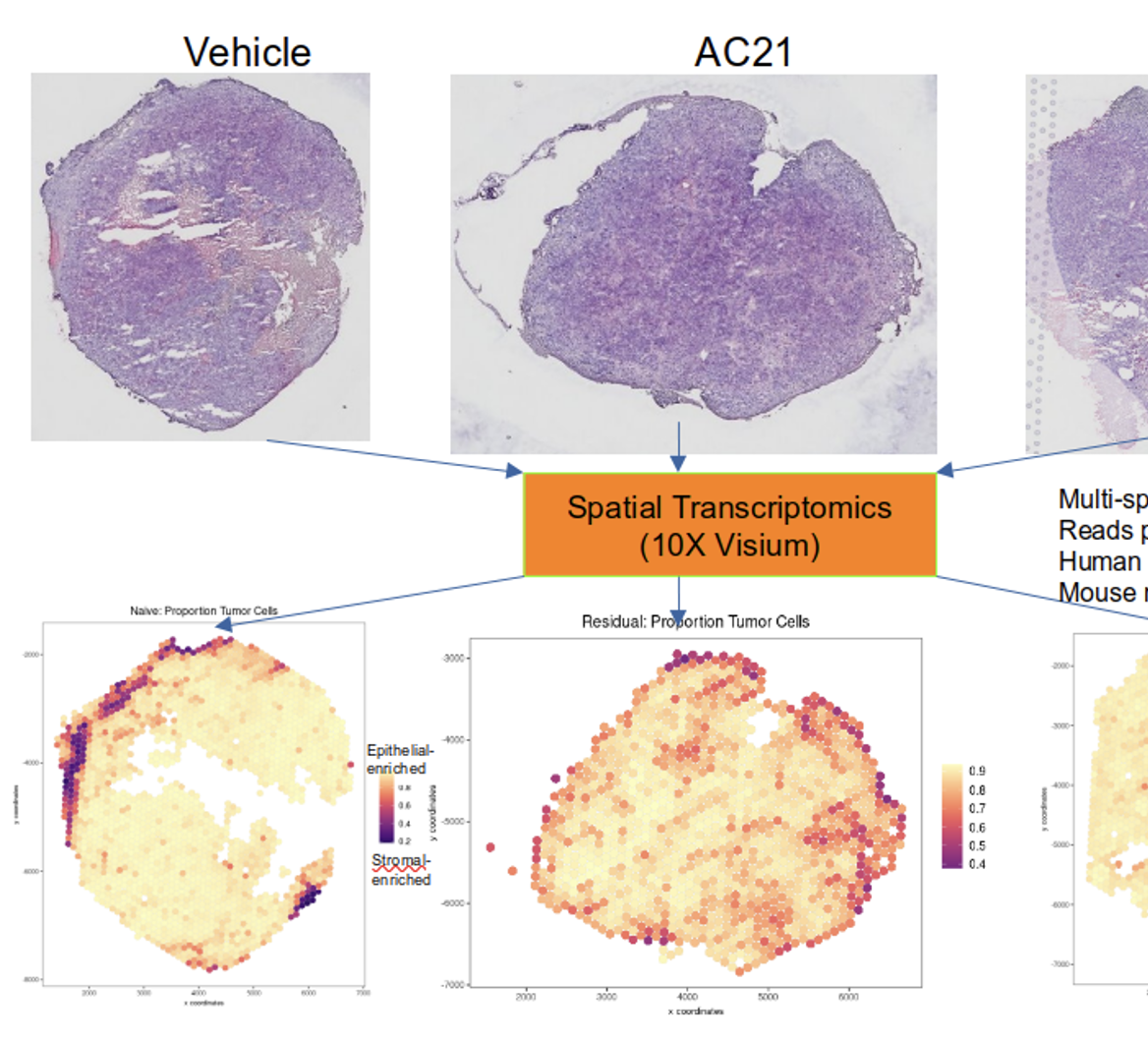
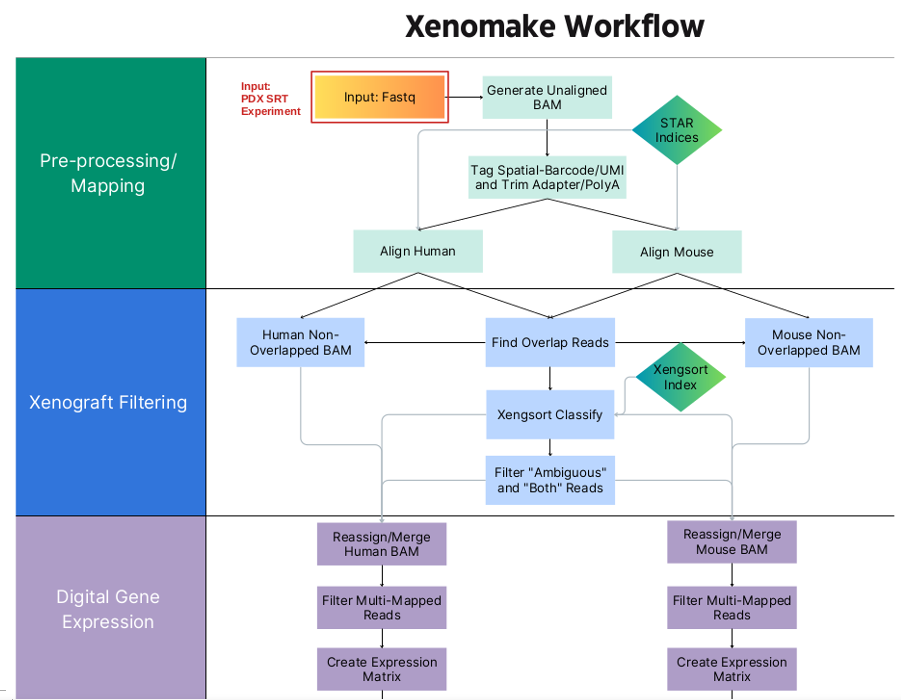
Welcome to the Zhu Lab
We are a computational biology and bioinformatics lab
focused on breast cancer research.
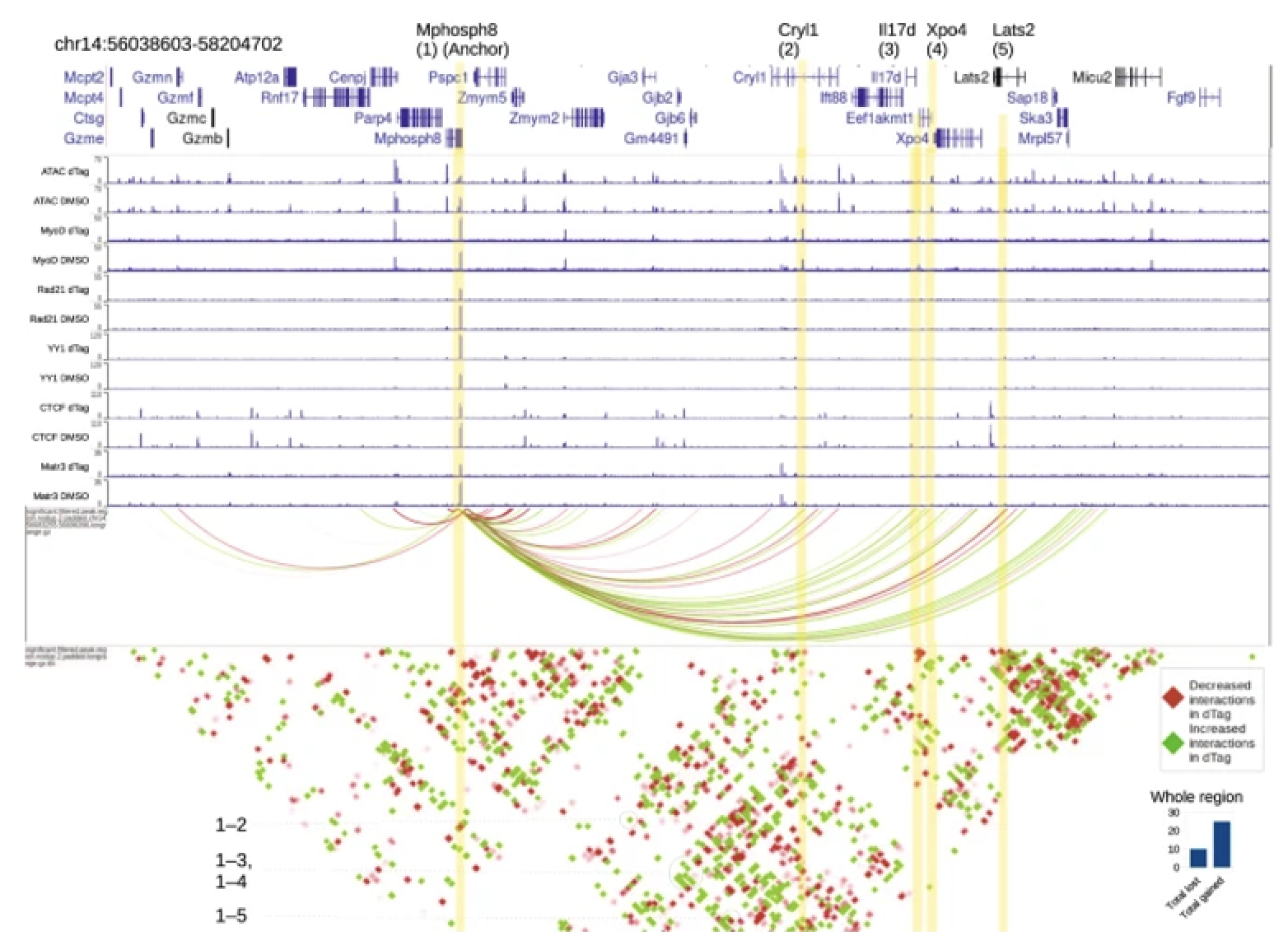
We employ
computational methods to study critical transitions of cancer

We use
multiomic data to identify predictive and prognostic biomarkers of cancer